Image Tab Tutorial#
Introduction#
This tutorial uses data in the file demo.hdf5. You can generate this file by following the instructions on Making the demo HDF5 file.
Open demo.hdf5 in hdf5view and select the dataset rgba. The hdf5view application window should look like this:
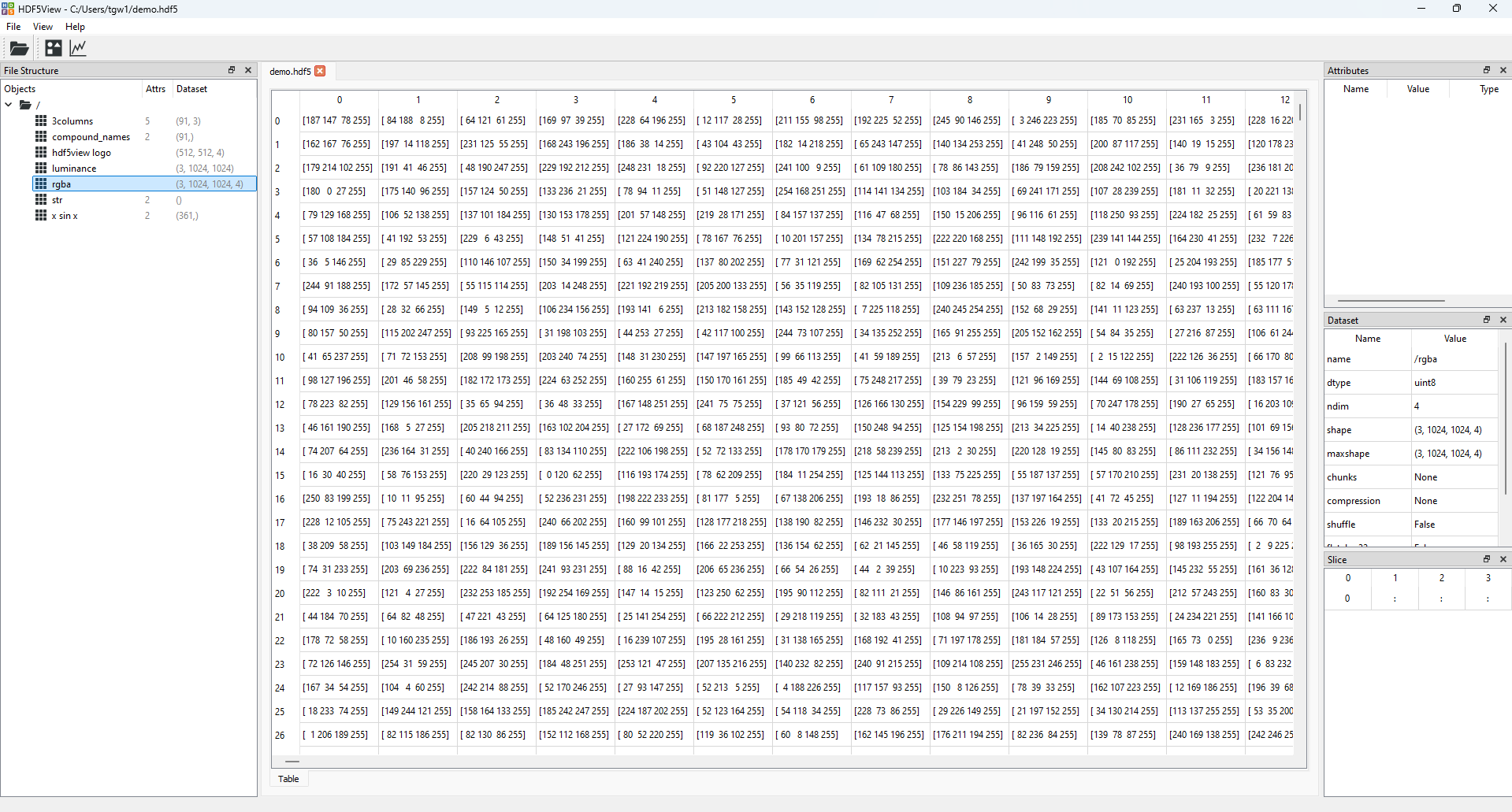
The state of the hdf5view application after selecting the dataset rgba in the File Structure of demo.hdf5.#
The dataset rgba has the shape (3, 1024, 1024, 4), which means that there are 3 rgba images stacked in the first axis and that the images are 1024 by 1024 pixels. Each pixel has four values associated with it: red, green, blue and alpha, which define the colour and transparency of the pixel.
To open an Image tab at the current dataset, click the image icon on the toolbar at the top of the application:
The image icon.#
The application will now look like this:
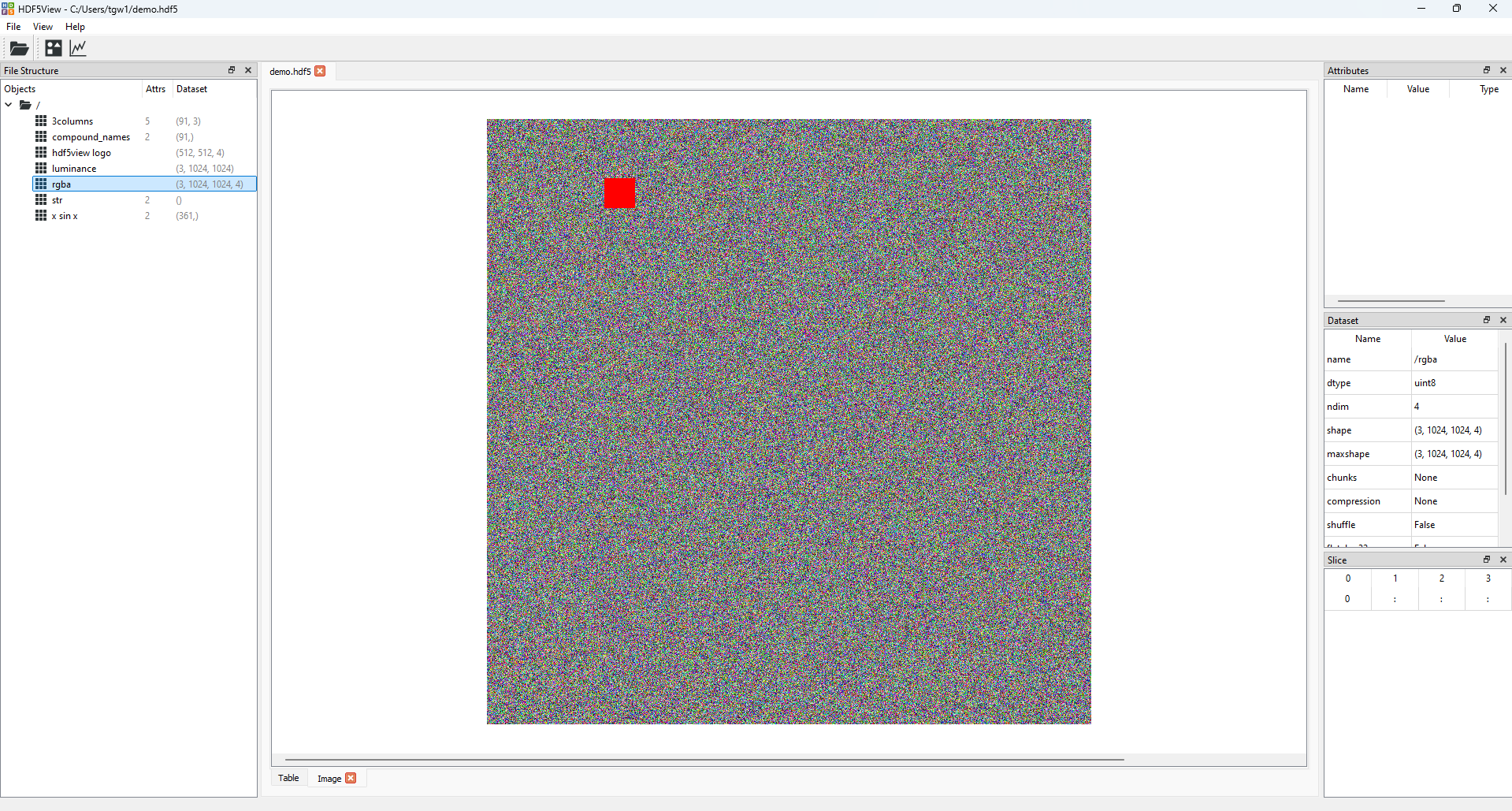
The state of the hdf5view application with an Image tab opened at the dataset rgba in demo.hdf5.#
In addition to the default Table tab, an Image tab is now open in the central tabbed area showing the image according to the default slice for this dataset, (0, :, :, :). This means that the top rgba image in the stack is being displayed.
You can have several
Imagetabs open at once.Imagetabs remember the dataset and slice if you switch to a different tab and back.Switching to a different dataset results in the default rendering behaviour for the image.
Using the Scroll Bar#
At the bottom of the Image tab, a scroll bar will appear if the dataset has more than 2 dimensions. Using the scroll bar, you can scroll through the first axis of the dataset. Taking the dataset rgba with the slice (0, :, :, :), we can move the scroll bar to the right. The image and slice will update accordingly and the new slice will be (1, :, :, :):
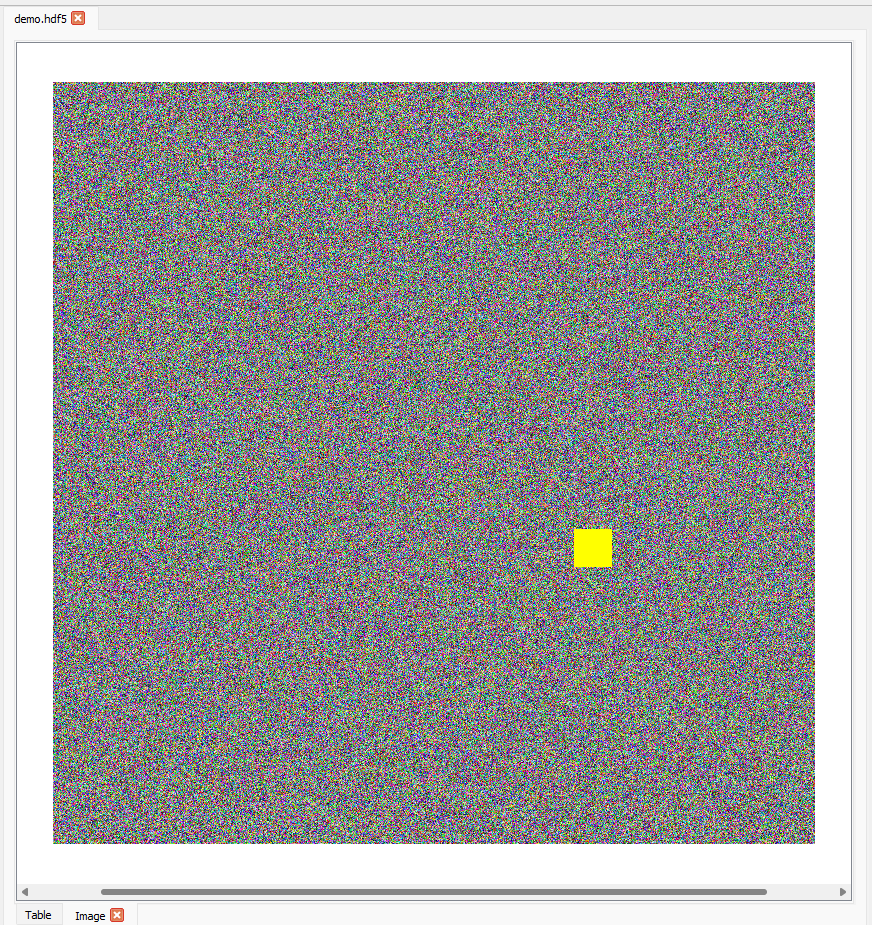
The Image tab having moved the scroll bar to the centre to select the second image in the stack of three in the dataset rgba in demo.hdf5.#
Slicing Images#
The Slice table can be used to select part of the image for a more detailed view. For example, if we want to select the region of the top image containing the red square in the dataset rgba, we can change the default slice (0, :, :, :) to (0, 72:174, 167:287, :):
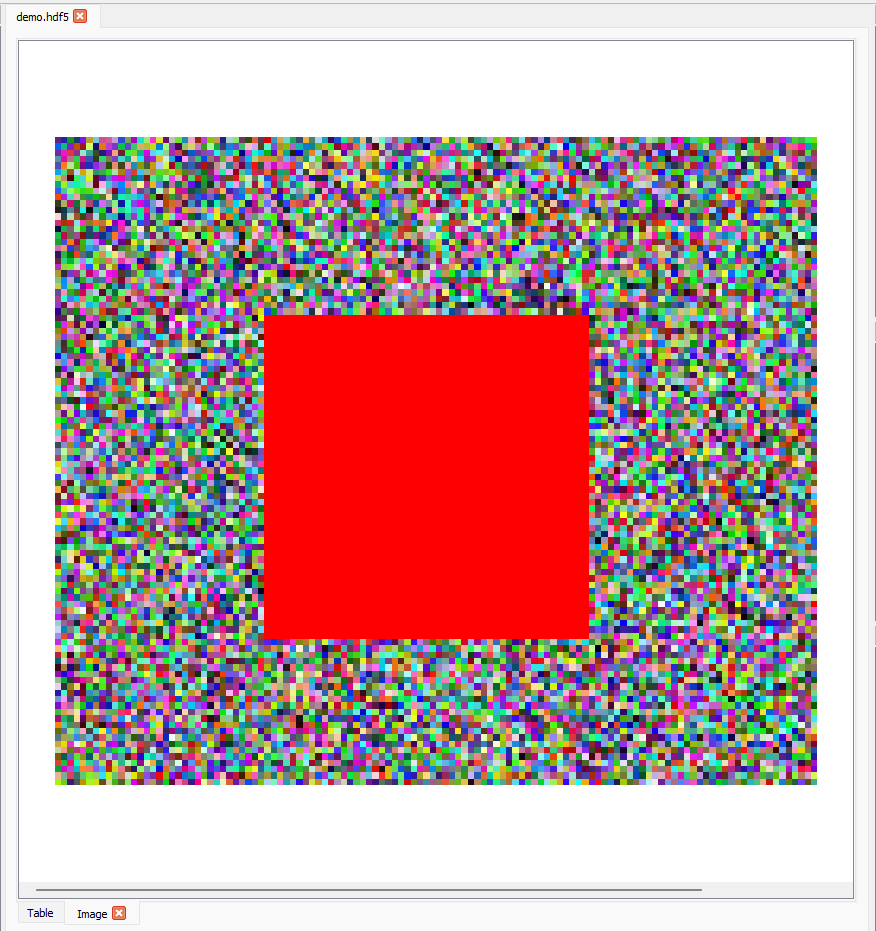
If we want to display the red channel data of this region as a luminance image, we can set the slice to (0, 72:174, 167:287, 0):
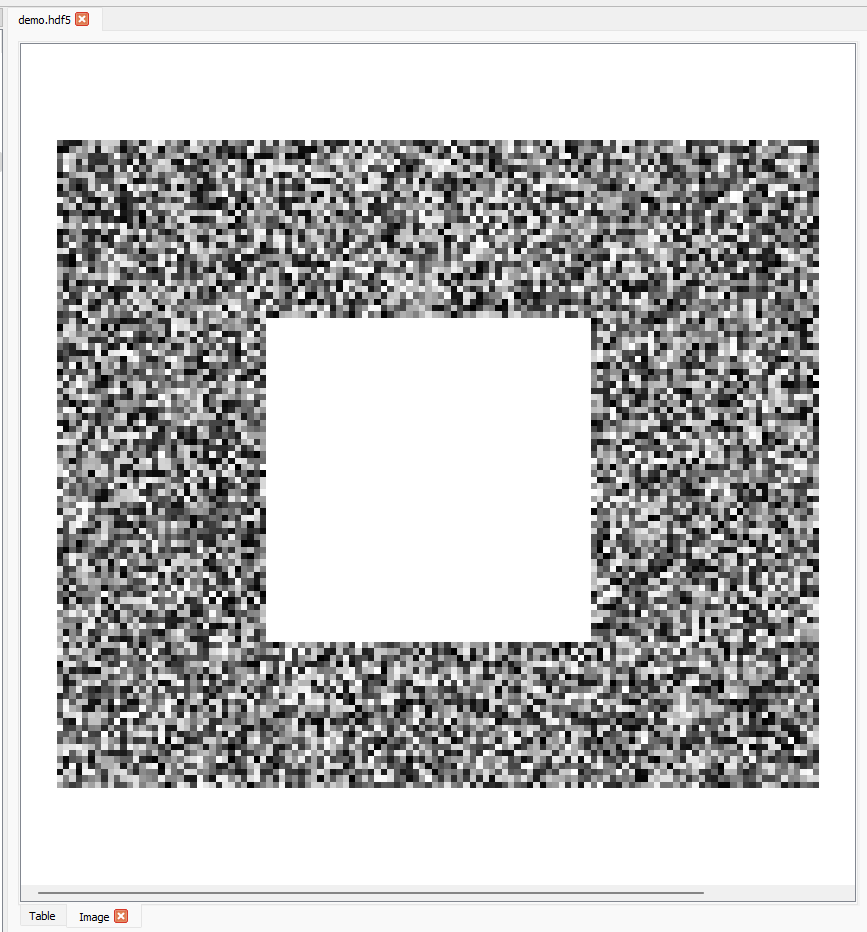
Or to display the green channel data as a luminance image, (0, 72:174, 167:287, 1):
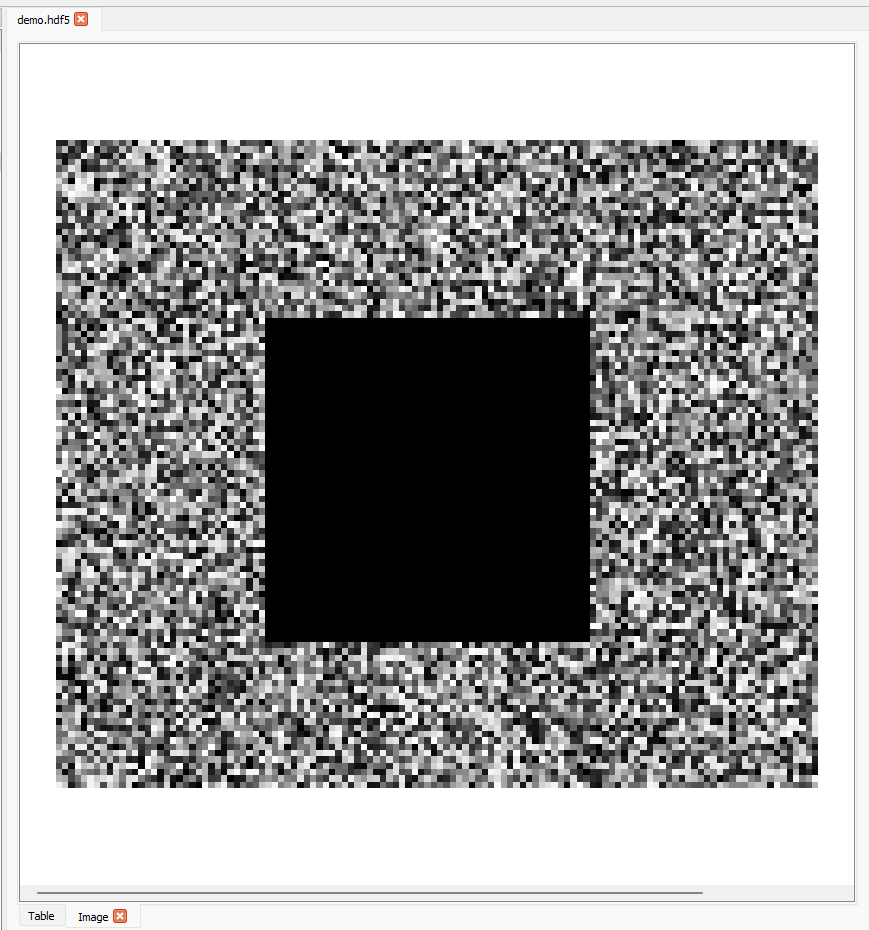
The scroll bar can be used to scroll through the first axis of the dataset and the slice on the other axes stays the same.
Interacting with Images#
When the mouse is inside the axes on an
Imagetab, the x and y coordinates of the mouse in pixel coordinates and the value of the pixel (either a single luminance value, 3 values for rgb or 4 values for rgba) will be shown on the message bar on the bottom left of the hdf5view application window.If you press and hold the left mouse button, you can drag the mouse to define a rectangle on an image. When you release the mouse button, the image will zoom to the rectangle selected.
To reset the zoom, right-click on the plot and select
View Allfrom the menu that appears.
Note
Using the rectangle select zoom does not change the Slice table in the current version.
The right-click menu contains several options for interacting with the image:
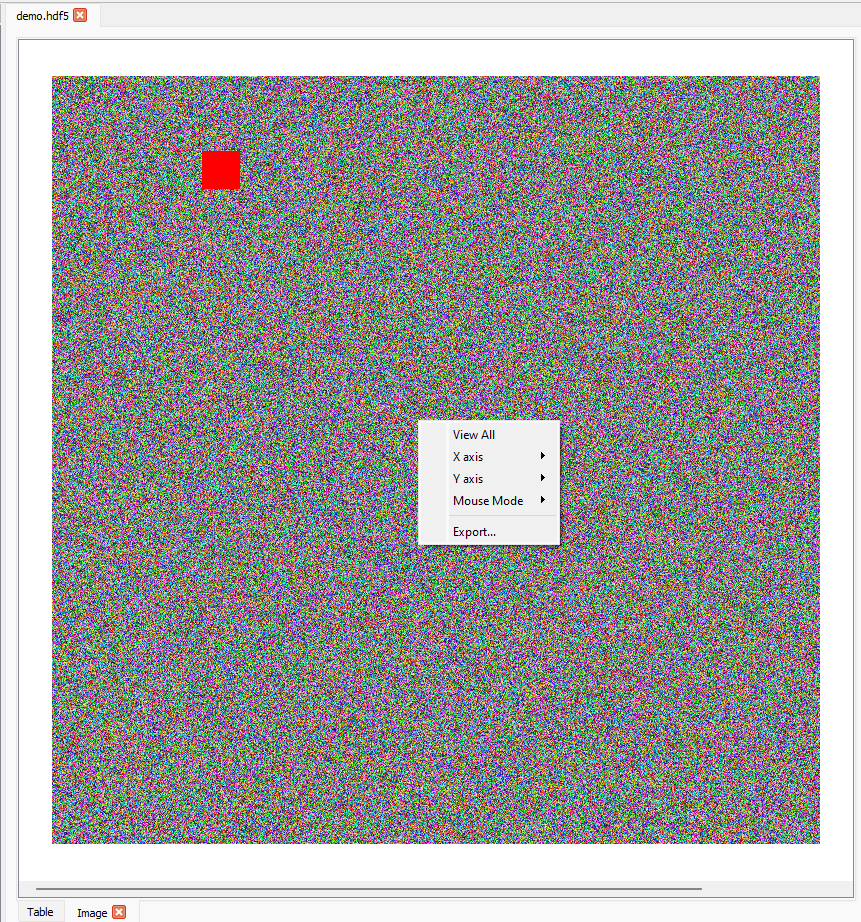
The
X axisandY axismenus provide options for manually scaling the axes.The
Mouse Modemenu enables switching between the default1 buttonmode (i.e. rectangle zoom, described above) and3 buttonmode where the left mouse button can be used to pan across the image, the mouse wheel zooms in or out, and the right mouse button can be used to stretch or compress an axis.
Exporting Images#
Images can be exported to several formats for further use. To access this option, right-click on an image and select Export.... The export wizard will then be shown. The export formats avilable are:
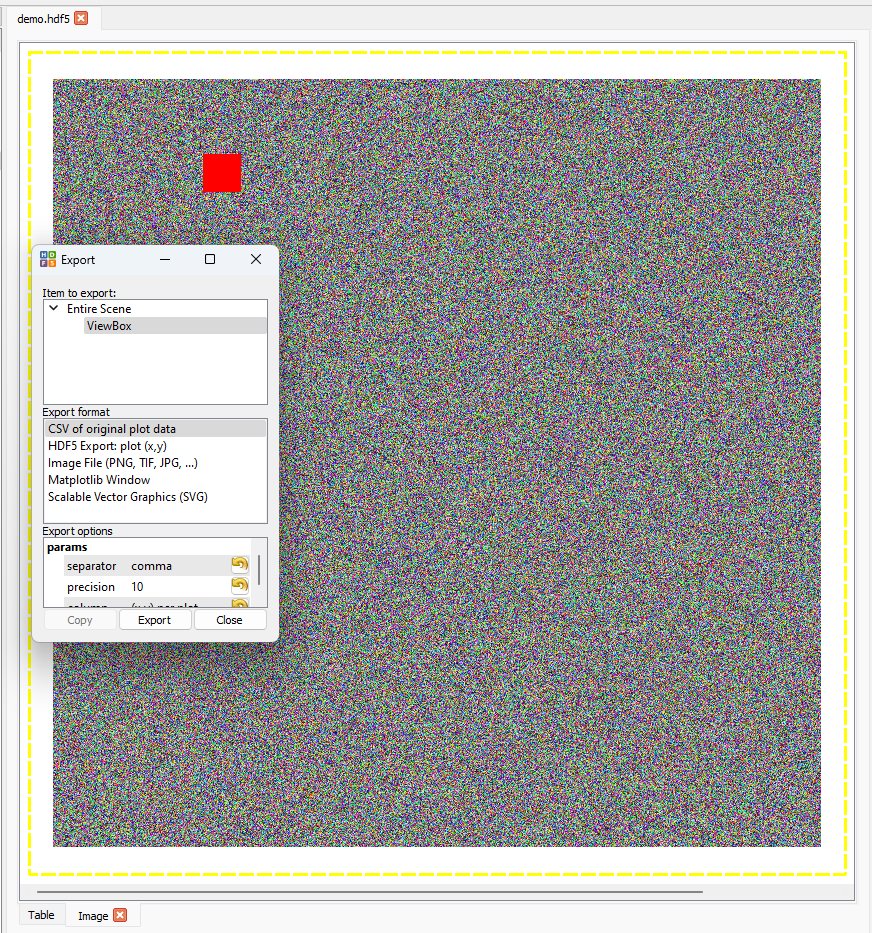
For further information, see the pyqtgraph documentation.
Default Slices on the Image Tab#
On selecting a dataset in the File Structure table, the Slice table adopts certain default values depending on the shape of the dataset. These default values are intended to act as a sensible starting point for displaying data in the Image tab. You can then change the Slice as needed.
The defaults on the Image tab for various dataset shapes are as follows:
ndim < 2: no image display possible
ndim = 2: all the data are plotted as a luminance (greyscale) image, Slice:
:, :ndim > 2 and shape[-1] in [3, 4]: here we assume that the data are rgb(a) images and plot as default the all data in the last 3 axes of the dataset for the first entry in any other axes, last 3 axes of Slice:
:, :, :.ndim > 2 and shape[-1] Not in [3, 4]: we plot luminance images as default showing all the data in the last 2 axes of the dataset for the first entry of any other axes, last 2 axes of Slice
:, :.
The default slices on the Image tab can be summarised as follows:
ndim |
condition |
interpretation |
default slice |
|---|---|---|---|
0 |
N.A., no image possible |
||
1 |
N.A., no image possible |
||
compound names |
N.A., no image possible |
||
2 |
luminance image |
(:, :) |
|
> 2 |
shape[-1] in [3, 4] |
rgb or rgba image(s) |
([0] * (ndim - 3) + [:, :, :]) |
shape[-1] Not in [3, 4] |
luminance images |
([0] * (ndim - 2) + [:, :]) |
You can test these default values by selecting different datasets on a Image tab in the file demo.hdf.

